Developing Scalable Feature Engineering DAGs
Our aim is to help your team think through the structure of your next data science project to ensure your development velocity does not slow down with the increasing age of your project. In this post we provide examples of how you can set your project up for long-term success. To this end we collaborated with Stefan Krawczyk from Stitch Fix to demonstrate how you can use two open-source tools, Hamilton and Metaflow, together to create and execute robust feature transformation pipelines as a part of ML workflows by leveraging directed acyclic graphs (DAGs). A great aspect of using these tools is that they are interoperable without need for a dedicated integration.
Successful collaboration in data science projects
Consider managing an important tabular dataset that several people are regularly reading and writing new features to. A common workflow in such a setting is to iterate on many feature transformation and downstream model building steps before selecting a satisfactory model.
As the project evolves and more team members join
- How do you manage the feature transformation history?
- How do you ensure modifications don’t change feature behavior inadvertently?
- How do you know what data/feature is related to what?
- How do you version your machine learning workflow metadata and dependencies?
- How does the team scale workflow steps by accessing necessary compute resources?
To solve the feature transformation issues listed above, encapsulating dataframe transformations in functions is an obvious way to modularize a code base. However, this doesn’t address important questions that don’t look important when first getting started, but matter when the code grows and changes hands like:
- What should you pass as input to and return from functions?
- Do you pass the whole dataframe? Do you pass specific columns of the dataframe?
- How are feature transformations related?
Moreover, data scientists know how important iteration is across steps in their workflows and that there is often much debugging to be done. As you iterate:
- How do you avoid recomputing work you have already done?
- If you only want a subset of features from the raw data, how do you avoid unnecessary feature transformations without re-writing your transformation code?
- If a workflow step fails, do you recompute everything prior to that step?
- How do you debug what steps in the workflow failed?
- How do you capture the state of the environment where failures occurred?
Another consideration is that as the number of transformations in a codebase grows overtime, determining feature dependencies by inspecting the codebase becomes more onerous. This makes discovering relationships and lineages through visualizations more valuable.
In data science projects, outcomes like unnecessary computation, code base complexity, and lack of discoverability can become significant sources of friction for developers. This is a problem if it blocks data scientists too much from focusing on levels of the stack where they contribute the most or have the most leverage.
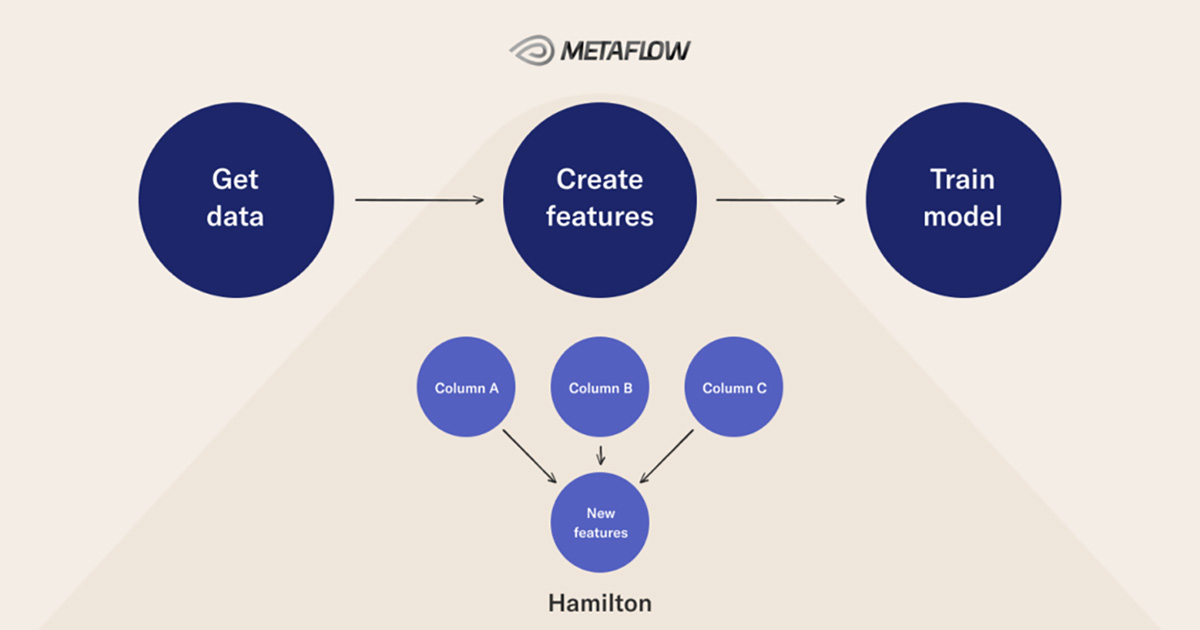
This post highlights DAGs as a useful way to structure data science workflows through an example using Hamilton and Metaflow. Both tools ask you to structure data science code as DAGs.
To read more about how these benefits emerge, check out: Why Should I Care About DAGs and Workflows in Data Science?
Intro to Hamilton
Hamilton is a micro-framework that helps you easily express and maintain feature transformation code as a DAG. It came from Stitch Fix and helps the Forecasting and Estimation of Demand (FED) team and their codebase stay productive. Managing a pandas code base of feature transforms over a few years usually isn’t a pleasant experience, but with Hamilton, you’ll avoid most of the pitfalls, and it’ll become an enjoyable experience, no matter the number of features you add, or who writes them!
Specifically, Hamilton forces data scientists to write Python functions that declare what they depend on via function parameters, and the function name specifies what the output of the function is.
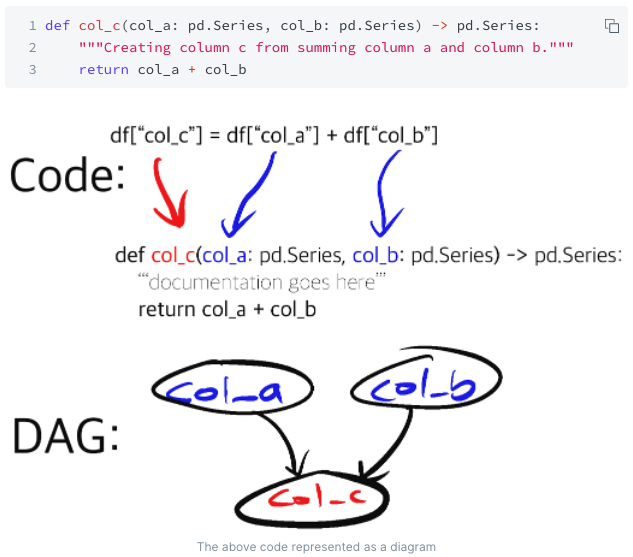
From this, the Hamilton library can build a DAG. With a DAG, which you get by instantiating a Hamilton Driver, then you just need to declare:
- the outputs you want
- the right feature names as input
and Hamilton will execute only the functions that are required to produce the outputs.
A guiding example
You can find the code example mentioned in the rest of this post in this repository. The example workflow includes: using Hamilton for feature transformation, several feature importance computations, and Metaflow for modeling steps, metadata tracking and workflow orchestration. You can find an introduction to Metaflow here.
The learning task in the example is multiclass classification and is defined in the paper: Prediction of absenteeism at work with a multinomial logistic regression model. The dataset was originally curated in the paper Application of a neuro fuzzy network in prediction of absenteeism at work. The original dataset has 20 features and labels encoded as integers.
Following the first paper in the previous paragraph our example encodes three label buckets:
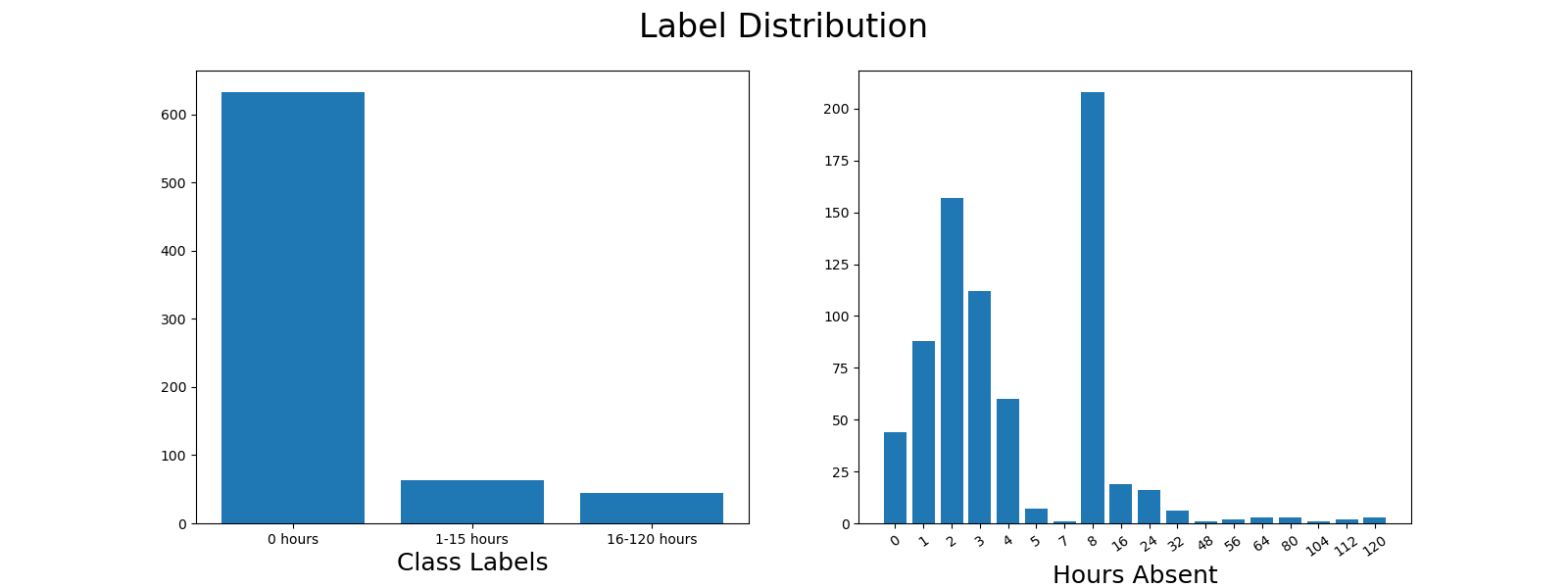
Our code example applies several feature importance methods and scikit-learn classifiers. For modeling steps, we track classifier performance on the following metrics: accuracy, macro-weighted precision, macro-weighted recall, macro-weighted f1, training time, and prediction time.
The remainder of this post describes how DAGs implemented in Hamilton and Metaflow can structure a workflow.
DAGs inside DAGs with Hamilton and Metaflow
Like Hamilton, Metaflow asks data scientists to structure code in DAGs. In Metaflow this is referred to as steps in a flow. In the code example guiding this post the flow leverages the following Metaflow features:
- branching and looping in the Metaflow DAG
- running arbitrary Python code – such as Hamilton feature transformation DAGs – in tasks
- dependency management with Metaflow’s conda integration
- cards to visualize artifacts produced by steps
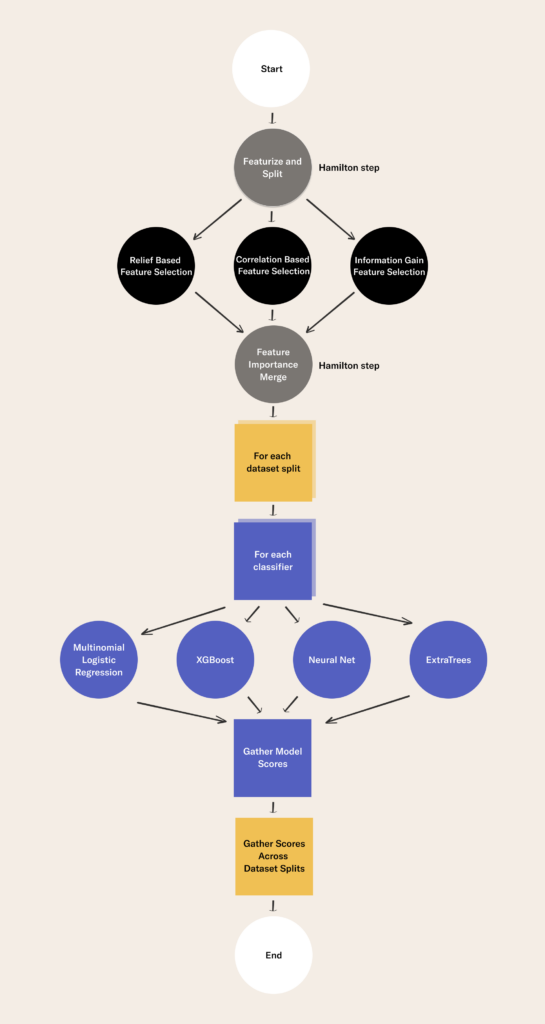
Applying Hamilton DAGs in our example
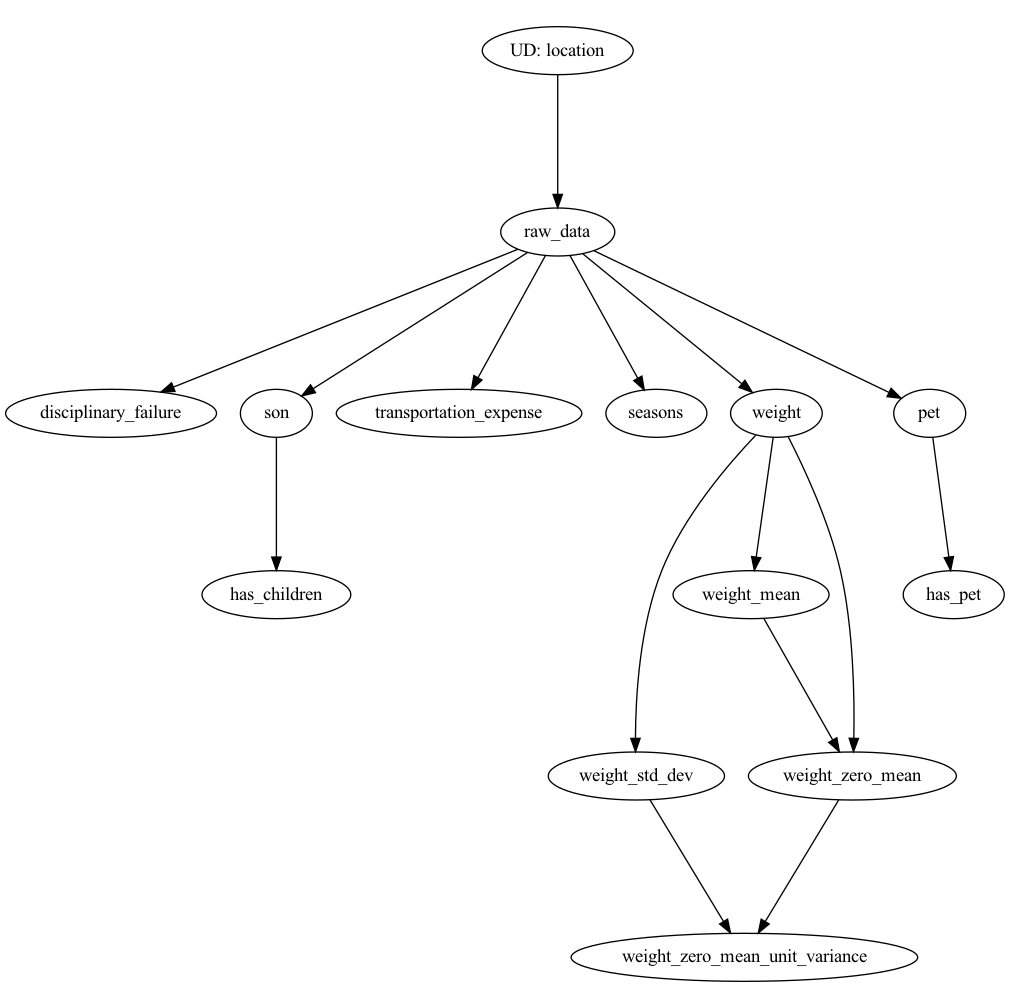
In our example we have created several modules that house our Hamilton functions:
data_loader.py– contains functions to load datanormalized_features.py– contains all numeric featuresnormalized_features_condensed.py– identical to normalized_features, but shows you some superuser ways to express Hamilton functions. See comments in the file for why or why not to use this approach.feature_logic.py– contains the rest of our features.
The numeric feature transformation functions are defined in normalized_features.py. For example, one function in this file extracts the mean age of people represented in data records:
def age_mean(age: pd.Series) -> float:
"""Average of age"""
return age.mean()The rest of the feature transformation functions are defined in feature_logic.py. For example, one function in this file extracts week days:
@extract_columns('day_of_the_week_2',
'day_of_the_week_3',
'day_of_the_week_4',
'day_of_the_week_5',
'day_of_the_week_6')
def day_of_week_encoder(
day_of_the_week: pd.Series) -> pd.DataFrame:
"""One hot encodes day of week into
five dimensions -
Saturday & Sunday weren't present.
1 - Sunday, 2 - Monday, 3 - Tuesday,
4 - Wednesday, 5 - Thursday,
6 - Friday, 7 - Saturday.
"""
return pd.get_dummies(day_of_the_week,
prefix='day_of_the_week')Hamilton’s @extract_columns decorator works on a function that outputs a dataframe that we want to extract the columns from and make them individually available for consumption. It expands a single function into many functions, each of which take in the output dataframe and output a specific column as named in the decorator.
To run it we just then instantiate a Hamilton Driver, passing it what it needs, and then executing it, requesting the features & outputs that we want back out.
In the example, we run a Hamilton Driver in two steps of a Metaflow flow. The flow is defined in the flow.py file. The two steps are featurize_and_split and feature_importance_merge. Hamilton DAGs are embedded in these steps of the Metaflow DAG. The Hamilton DAG nodes are features and edges are transformations. The Metaflow DAG nodes are more general workflow steps and edges that tell an orchestrator the order steps should be executed. With Hamilton we are modeling a specific aspect of the data science workflow, namely feature transformation. With Metaflow we model a macro view of the workflow(s) describing our project. Metaflow steps can contain anything you express in Python code.
The main thing to pay attention to is that in addition to the self-documenting, easy-to-read transformation code Hamilton enforces we are re-using the same code in both steps to request different feature subsets. In featurize_and_split we execute the following Driver code to select all features.
from hamilton import driver
import data_loader
import feature_logic
import normalized_features
dr = driver.Driver({
"location": Config.RAW_FEATURES_LOCATION},
data_loader,
feature_logic,
normalized_features)
features_wanted = [
n.name for n in dr.list_available_variables()
if n.name not in EXCLUDED_COLS and \
n.type == pd.Series]
full_featurized_data = dr.execute(features_wanted)

In the feature_importance_merge step we select only the top k features based on a feature selection policy applied to the outputs of earlier steps in the DAG that represents the flow.
Better together
We’ve explained the code, now let’s dive into the benefits.
Metaflow allows a macro overview of the model building process, Hamilton allows a micro view into a complex part of the model building process – creating features. Both are DAGs and guide users to structuring code bases appropriately.
Maintainable and naturally documented feature transformation history
Hamilton functions are documentation friendly and always unit testable. Metaflow has cards to visualize contents of a step and share them easily. In our example code, you should look at how readable the metaflow steps are and the Hamilton feature transform functions. Take a look at the readme for the guiding example to see how you can visualize steps with Metaflow cards.
Modular execution of feature transformation pipelines – only compute what you need to
Hamilton enables one to easily describe the superset of features and to only compute what is needed at your request. You can see this subset happening in the feature_importance_merge of the flow. Metaflow allows you to model different feature needs as different steps.
Insight into the relationship DAG between feature transformations
You can visualize the macro workflow with Metaflow. You can visualize the micro feature transformation workflow with Hamilton. Enabling both allows you to see end-to-end relationships, as well as interactions within a complex step. You can use Metaflow features to debug a failure in a task (steps become tasks at runtime) in the flow. Should your debugging lead you to a featurization step, you can then access the state and peer into the Hamilton DAG, where you can step through individual feature steps.
Built-in versioning of your machine learning workflows including dependency management
Metaflow captures the state of your flow runs including dependencies and any metadata you want to track. This enables you to resume flows from failed tasks and helps you share workflows in a more portable, production-ready form. On a more granular level, Hamilton enables feature versioning via Git and Python packaging.
DAGs to scale steps in your workflow to any compute resources
No one likes migrations. When dealing with large datasets and models, an extension to the flow template provided in this example is to have Metaflow steps run on remote machines. To do this, you can add Metaflow’s @batch or @kubernetes decorator above a step and specify how much processing power and memory the step requires. Alternatively you can send all steps to one of these compute environments by adding --with=batch or --with=kubernetes in the command line.
Since by default Hamilton expects dataframes it operates on to fit in memory, a convenient pattern for scaling Hamilton is to wrap the Metaflow step containing Hamilton driver execution with an aforementioned decorator for defining compute requirements. Another route is to leverage Hamilton’s ability to scale your dataframe transformations with Dask, Ray, or Pandas on Spark. Then you can scale the RAM required to support a Hamilton feature transformation run by changing the amount of memory requested for the step. Message us on slack if you have ideas for how you would design a system to scale Hamilton with Metaflow!
Summary
Given the experimental nature of their work, data scientists often need to prioritize iterating quickly on current tasks. However, in the long run, it can increase the speed of future iterations to focus on workflow structure at the beginning.
“Give me six hours to chop down a tree and I will spend the first four sharpening the axe.” – Abraham Lincoln
The benefits of structuring code projects well compound over time and across projects. Make sure to pick tools that help you structure projects, iterate on experiments, and move to production without adding headaches.
Check out these links to get started developing with Hamilton and Metaflow today!
Start building today
Join our office hours for a live demo! Whether you're curious about Outerbounds or have specific questions - nothing is off limits.










